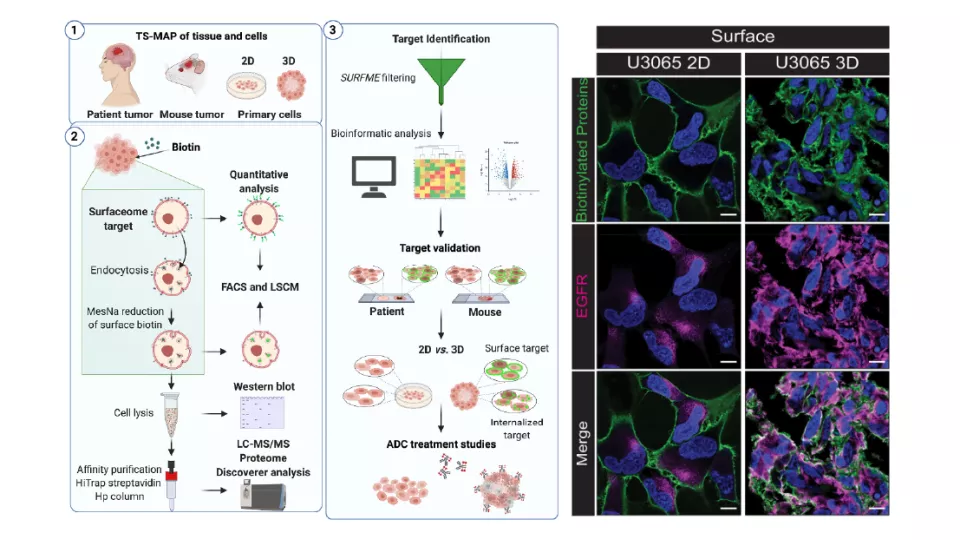
On February 2022 our group had a new paper released in the journal PNAS, titled “Landscape of Surfaceome and Endocytome in Human Glioma is Divergent and Depends on Cellular Spatial Organization”, which describes the TS-MAP protocol, a procedure for proteomics-based target identification aimed at a better understanding of how to harness the tumor surfaceome for personalized immunotherapy.
The study investigated how the protein presentation at surface, as well as its uptake, is affected by cellular spatial organization when comparing 2D versus 3D cultured glioma cells. Furthermore, taking the example of the well-known glioma target EGFR, the study shows that cellular spatial organization at the experimental level can drastically affect the results of early immunotherapy assays.
When applied to freshly-resected tumors, the protocol revealed that the surfaceome is highly divergent between sets of patients, independently from tumor grade or histology, and that the TS-MAP can be a valuable tool on the help to uncover potentially targetable proteins for personalized therapy development. This approach is highly versatile and can be widely applicable to a variety of solid tumors, or it can be implemented to provide useful insights into the design of adjuvant immunotherapies specifically targeted at resistance mechanisms or could be expanded for the purpose of noninvasive brain tumor diagnosis.